Storage and Files¶
Storage of data on Gencove Explorer relies on two main mechanisms:
- Local storage
- Cloud storage, aka EOS (Explorer Object Storage)
Each Gencove Explorer instance contains its own local storage as you would expect with any virtual machine. However, this SSD storage space is limited, and is intended to be a transient or intermediary store for your data.
Larger persistent data is intended to be stored on EOS cloud storage, which is private to your organization. To that end, we provide several mechanisms through the Explorer SDK and CLI to enable easily managing your data in the cloud.
Local Instance Storage¶
The directory /home/explorer is your personal local storage area. Any programs, scripts, or data
files you work with in Jupyter Lab can be saved here. To ensure optimal system performance, please
keep track of
your storage usage and manage your files appropriately, offloading any large data files to cloud
storage as necessary (described in the following section).
By default, each Explorer instance is allocated 200GB of local disk space.
Running the command df -h through the Explorer terminal, you will see output similar to the
following:
Filesystem Size Used Avail Use% Mounted on
overlay 196G 5.0G 181G 3% /
tmpfs 64M 0 64M 0% /dev
/dev/nvme1n1 196G 5.0G 181G 3% /home/explorer
shm 64M 0 64M 0% /dev/shm
/dev/nvme0n1p1 50G 2.6G 48G 6% /opt/ecs/metadata/111
tmpfs 7.7G 0 7.7G 0% /proc/acpi
tmpfs 7.7G 0 7.7G 0% /sys/firmware
While using the Gencove Explorer system, it is
important to regularly take note of how much local storage space you have used. This can easily
retrieved by running df -h /home/explorer in the terminal. If you are nearing your storage limit
of 200 GB, consider offloading larger data files to cloud storage, described in the
following section.
Cloud Storage, EOS¶
Data files commonly used in genomics applications are often very large (tens or even hundreds of Gb), and therefore can be unwieldy to work with.
Your Gencove Explorer instance and Analysis jobs are all configured with access to EOS (Explorer Object Storage) - private cloud object storage which can only be accessed by users in the Gencove Organization you belong to.
EOS is essentially a lightweight wrapper around AWS S3 object storage that aims to simplify access to the appropriate user, organization, and Gencove-wide locations on S3.
In addition to its primary storage capabilities, EOS also provides archive and restore functionalities. This feature allows users to manage their data lifecycle by archiving infrequently accessed data and restoring it when needed. This ensures optimal storage utilization and cost efficiency.
EOS locations¶
EOS URIs always start with e://. There are three top-level namespaces for EOS:
- User:
e://users/<user-id>/- read/write for current user, only read for the rest of your organization
e://users/me/is accepted as shorthand for the current user's<user-id>e://users/<user-email>is accepted in lieu of<user-id>e://users/will display<user-email>and<user-id>of all users in your organization
- Organization:
e://org/- read/write for entire organization
- Gencove:
e://gencove/- read for entire organization
EOS can be accessed using the Gencove Explorer CLI and Explorer SDK.
EOS via CLI¶
The Gencove CLI provides AWS CLI equivalents of ls, cp, rm, and sync commands that can be used for Explorer data management. Note that the AWS CLI is a dependency for this functionality. For example:
For ease of use, the gencove explorer data command is also available via the ged
alias. The equivalent ged command to the example above would be:
List, ls¶
Listing files can be accomplished with the ls command:
Copy, cp¶
Files can be uploaded and downloaded using the cp command:
Synchronize, sync¶
Files can be synced in bulk using the sync command:
Delete, rm¶
Remote files can be deleted using the rm command:
Archive, archive¶
Archive files in bulk using the archive command:
Archive files individually:
NOTE: Archived files reduce storage costs, but need to be restored before accessing them.
Restore, restore¶
Restore archived files in bulk using the restore command:
Restore files individually:
¶
EOS via SDK File object¶
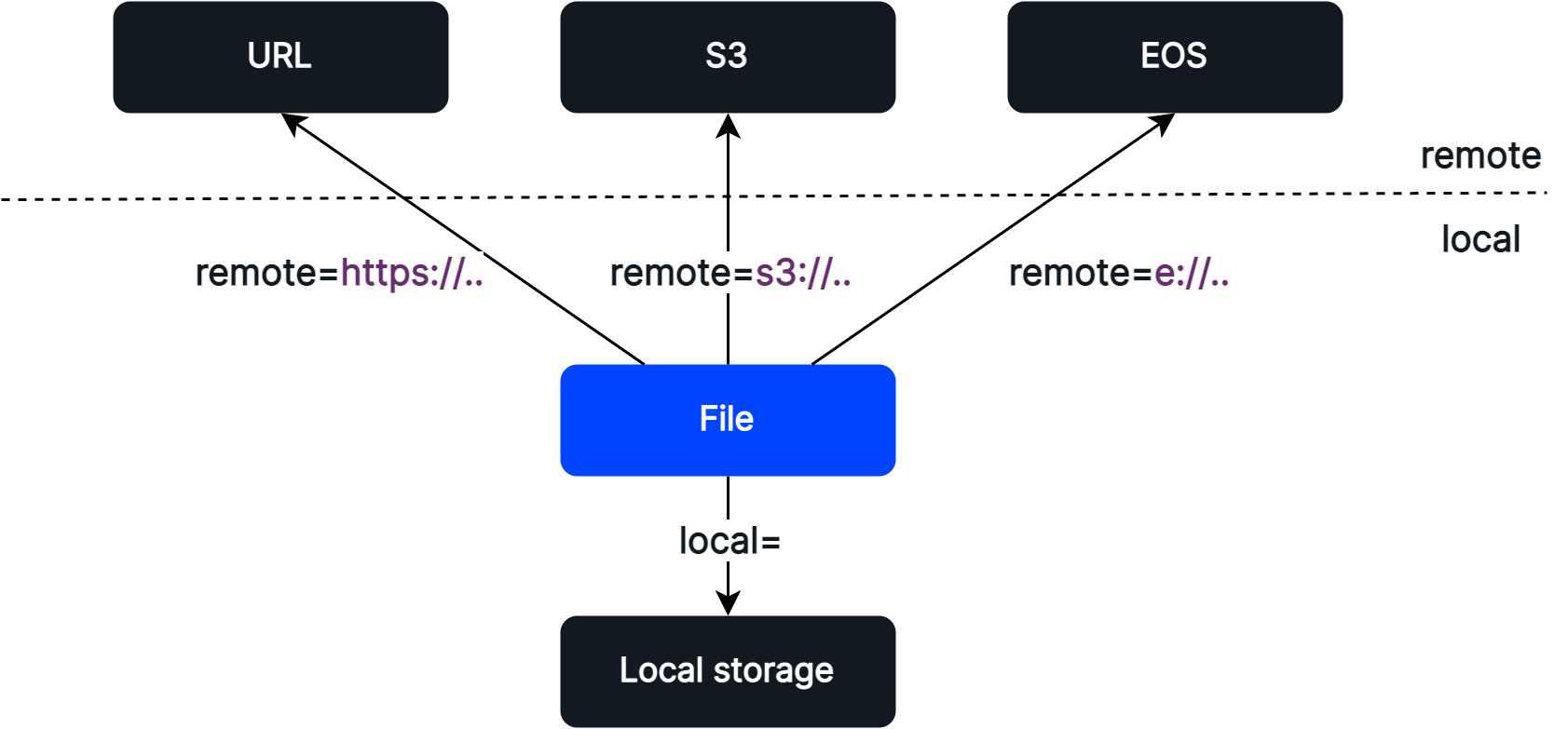
The Explorer SDK comes with an inbuilt abstraction of a File object, which represents a file with a local and/or remote location. It provides a way to specify and transfer (download and upload) files between local and remote storage.
Neither the local nor remote locations need to exist when the object is created, which effectively allows working with files in a "lazy" manner. This way, the abstraction of a file can be worked with but the actual upload or download of a potentially very large file is not effected until an explicit method call is invoked.
Neither local nor remote location need to be specified for the File object. If left unspecified, the following defaults are assumed:
- Local: a random filename in the
/tmpdirectory - Remote: a random key in a temporary EOS location
In this section, we describe the File object at a high level, and describe the various methods available to access and transfer (download and upload) remote files.
💡 Note that “local” in this section refers to your Explorer Instance storage or Explorer Analysis job storage.
Parameters¶
Parameter remote
The remote parameter allows you to specify the remote location of the file. The remote parameter supports a number of object types, including:
S3FileEFileURLFileNamedFile- Plain strings
When a string is supplied to remote, the SDK will do a best attempt at identifying the proper object to map to your remote parameter. For example:
from gencove_explorer.file import File
f = File(remote="e://users/me/example_e.txt")
print(f"{f.remote=}") # f.remote=EFile(path='e://users/me/example_e.txt')
The above demonstrates how the e:// prefix is used to infer the EFile object type. Likewise, similar logic applies to s3:// and https:// prefixes, which will create S3File and URLFile object types, respectively.
In addition, supported objects can be passed in directly. For example:
from gencove_explorer.file import File, EFile
f = File(remote=EFile(path="e://users/me/example_e.txt"))
The NamedFile type provides a shorthand for EOS storage and behaves similarly to an EFile. When none of the known protocols are matched (e.g. https://, e://, s3://) in the remote string, the system will assume a NamedFile. For example, the following two File objects are equivalent:
from gencove_explorer.file import File
f1 = File(remote="e://users/me/example_e.txt") # <- produces EFile
f2 = File(remote="example_e.txt") # <- produces NamedFile
Parameter local
The local parameter allows you to specify the local location of a file. The parameter represents either an existing file, or a desired location for a file to be downloaded to, depending on context.
The following example demonstrates the following behavior involving the local parameter:
- Uploading a local file to EOS
- Downloading a remote file to a local location
from gencove_explorer.file import File
from pathlib import Path
import sh # import necessary package
# Create a dummy file to upload
upload_demo_file = Path("/tmp/demo.txt")
sh.echo("example", _out=upload_demo_file)
# Upload local file to remote destination on EOS
f1 = File(local=upload_demo_file, remote="upload_demo/upload_demo_file.txt")
f1.upload()
# Specify local path when creating the File object
f2 = File(remote="upload_demo/upload_demo_file.txt", local="/tmp/example_download.txt")
f2.download()
# Alternatively local path when calling .download()
f3 = File(remote="upload_demo/upload_demo_file.txt")
f3.download(local="/tmp/example_download_2.txt")
# Print path to local copy of files
print(f2.local)
print(f3.local)
Additional information on the download() and upload() methods can be found in the following sections.
💡 All parameters `of theFileobject must be provided by keyword, otherwise, the exceptionFile.init() takes 1 positional argument but 2 were given` will be raised.
Optional parameters: auto_archive, auto_expire
By default auto_archive is True. Which means remote files are automatically archived after 10 days. To disable this feature create files with File(remote=..., auto_archive=False). After the file is archived it can be recovered by calling file.restore().
By default auto_expire is False. It can be enabled with File(remote=..., auto_expire=True), in that case the file will be automatically deleted after 7 days. There's no way to recover the file after that.
Downloading files¶
Once a File object that refers to a remote file is created, you can retrieve a local copy via the download() method. For example:
from gencove_explorer.file import File
# Copy file from EOS via remote
f1 = File(remote="e://users/me/example_e.txt")
f1.download()
# Copy file from EOS via name
f2 = File(remote="example_e.txt")
f2.download()
# Print path to local copies of files
print(f1.local)
print(f2.local)
Other remote objects
For files that exist remotely outside of EOS, you can use the remote parameter of File. The File object supports a number of sources, including:
- S3 paths
- URLs
Downloading an S3 object
from gencove_explorer.file import File
f3 = File(remote="s3://bucket/path/file.txt")
# Copy file from S3 (you must have the necessary IAM permissions)
f3.download()
# Print path to local copy of file
print(f3.local)
Downloading a file from a URL
For files that exist on the public Internet, you can provide the URL via the remote parameter to File:
from gencove_explorer.file import File
f4 = File(remote="https://...")
# Copy file from URL (URL must be publicly accessible)
f4.download()
# Print path to local copy of file
print(f4.local)
💡 Note that downloading files from FTP links via File.download() is not currently supported.
Uploading files¶
Files can also be uploaded from local storage to EOS (or S3 more generally):
from gencove_explorer.file import File
import sh # import necessary package
f1 = File(
local="~/file.txt",
remote="e://users/me/project-1/file.txt"
)
sh.echo("File content", _out=f1.local)
f1.upload()
Executing files¶
File objects can be executed as scripts by using the execute() method:
from gencove_explorer.file import File
import sh # import necessary package
f = File(local="~/script.sh")
sh.echo("#!/bin/sh\necho Hello World", _out=f.local)
f.execute()
Command-line parameters can be passed to execute() as follows:
from gencove_explorer.file import File
import sh # import necessary package
f = File(local="~/script.sh")
sh.echo("#!/bin/sh\necho $1 $2", _out=f.local)
f.execute("parameter1", "parameter2") # equivalent to: ~/script.sh parameter1 parameter2
In case it is preferable to process the output instead of printing it to the terminal, execute() can be configured to provide the output via its return value by setting capture_output to True:
from gencove_explorer.file import File
f = File(local="~/script.sh")
c = f.execute(capture_output=True)
print(c.stdout) # Standard output
print(c.stderr) # Standard error
File.execute() uses /bin/sh as the default interpreter, but any interpreter can be specified by setting the appropriate shebang line in the file. For example, to use Python as the interpreter add the following line to the top of the Python script:
Finally, a real-world example for executing a shell script (~/script.sh) on an array of inputs utilizing the features described in this section:
from gencove_explorer.file import File
from gencove_explorer.analysis import Analysis, InputShared
a = Analysis(
input=["a","b","c"],
input_shared=InputShared(
script=File(local="~/script.sh").upload()
),
function=lambda ac: ac.input_shared.script.execute(ac.input),
).run()
Temporary files¶
It is also possible to use the File object for creating temporary local and/or remote files.
💡 Temporary files have an automatically generated filename that is guaranteed to be unique. The user should not expect that these files are permanently stored. We recommend using them for intermediate analysis results, while named files should be used for inputs and outputs.
To create a temporary remote file, exclude the name and remote parameters:
from gencove_explorer.file import File
# Create File object with no explicit destination
f = File(local="~/file.txt")
# Upload to temporary location
f.upload()
To create a temporary local file, exclude the local parameter:
from gencove_explorer.file import File
import sh # import necessary package
# Create File object with explicit destination
f = File(remote="e://users/me/project-1/123.txt")
sh.echo("123", _out=f.local)
# Upload to temporary location
f.upload()
To create a file that is temporary both locally and remotely, exclude all parameters:
from gencove_explorer.file import File
import sh # import necessary package
# Create temporary File object
f = File()
sh.echo("123", _out=f.local)
# Upload to temporary location
f.upload()
# Get destination
print(f.remote)
Generating URLs for remote File objects¶
It is possible to generate a temporarily accessible URL for a file in EOS via the remote url property.
💡 The URLs generated with this method:
- Provide access to the file over the public Internet by anyone who has the URL
- Expire after 48 hours
Below is an end-to-end example where we copy a local file to EOS, then obtain a URL for it.
from gencove_explorer.file import File
import sh # import necessary package
# Create File object with no explicit destination
f = File(local="~/file.txt")
# Write contents to file
sh.echo("URL example", _out=f.local)
# Upload to temporary location
f.upload()
# Generate and print temporary URL for file
print(f.remote.url)
Sharing files within your organization¶
Files can be easily shared within your organization by sharing the EOS URI (e://...) of objects.
💡 Files created in your user namespace (e://users/me/) need to be made "shareable" by replacing the me shorthand with your user id. To simplify this process, the Explorer SDK provides a convenient File object attribute named path_e_shareable.
from gencove_explorer.file import File
# Create File object in user namespace
f = File(local="~/file.txt", remote="e://users/me/project-1/file.txt")
# Copy to temporary location on EOR
f.upload()
print(f.path_e_shareable)
EOS URIs of files created in the organization namespace (e://org/) can be shared within your organization without modification.